Ribose-5-phosphate isomerase (Rpi) is an enzyme that catalyzes the conversion between ribose-5-phosphate (R5P) and ribulose-5-phosphate (Ru5P). It is a member or a larger class of isomerases which catalyze the interconversion of chemical isomers (in this case structural isomers of pentose). It plays a vital role in biochemical metabolism in both the pentose phosphate pathway and the Calvin cycle. The systematic name of this enzyme class is D-ribose-5-phosphate aldose-ketose-isomerase.
Structure

Rpi exists as two distinct proteins forms, termed RpiA and RpiB. Although RpiA and RpiB catalyze the same reaction, they show no sequence or overall structural homology.2 According to Jung et al., an assessment of RpiA using SDS-PAGE shows that the enzyme is a homodimer of 25 kDa subunits. The molecular weight of the RpiA dimer was found to be 49 kDa by gel filtration. Recently, the crystal structure of RpiA was determined. (please see http://www3.interscience.wiley.com/cgi-bin/fulltext/97516673/PDFSTAR)
Due to its role in the pentose phosphate pathway and the Calvin cycle, RpiA is highly conserved in most organisms, such as bacteria, plants, and animals. RpiA plays an essential role in the metabolism of plants and animals, as it is involved in the Calvin cycle which takes place in plants, and the pentose phosphate pathway which takes place in plants as well as animals.
All orthologs of the enzyme maintain an asymmetric tetramer quaternary structure with a cleft containing the active site. Each subunit consists of a five stranded β-sheet. These β-sheets are surrounded on both sides by α-helices. This αβα motiff is not uncommon in other proteins, suggesting possible homology with other enzymes. The separate molecules of the enzyme are held together by highly polar contacts on the external surfaces of the monomers. It is presumed that the active site is located where multiple β-sheet C termini come together in the enzymatic cleft. This cleft is capable of closing upon recognition of the phosphate on the pentose (or an appropriate phosphate inhibitor). The active site is known to contain conserved residues equivalent to the E. coli resides Asp81, Asp84, and Lys94. These are directly directly involved in catalysis.
Mechanism
In the reaction, the overall consequence is the movement of a carbonyl group from carbon number 1 to carbon number 2; this is achieved by the reaction going through an enediol intermediate (Figure 1). Through site-directed mutagenesis, Asp87 of spinach RpiA was suggested to play the role of a general base in the interconversion of R5P to Ru5P.
The first step in the catalysis is the docking of the pentose into the active site in the enzymatic cleft, followed by allosteric closing of the cleft. The enzyme is capable of bonding with the open-chain or ring form of the sugar-phosphate. If it does bind the furanose ring, it next opens the ring. Then the enzyme forms the eneldiol which is stabilized by an lysine or arginine residue. Calculations have demonstrated that this stabilization is the most significant contributor to the overall catalytic activity of this isomerase and a number of other like it.
Metabolic role
Pentose phosphate pathway
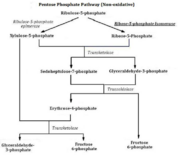
In the non-oxidative part of the pentose phosphate pathway, RpiA converts Ru5P to R5P which then is converted by ribulose-phosphate 3-epimerase to xylulose-5-phosphate (figure 3). The end result of the reaction essentially is the conversion of the pentose phosphates to intermediates used in the glycolytic pathway. In the oxidative part of the pentose phosphate pathway, RpiA converts Ru5P to the final product, R5P through the isomerization reaction (figure 3). The oxidative branch of the pathway is a major source for NADPH which is needed for biosynthetic reactions and protection against reactive oxygen species.
Calvin cycle
In the Calvin cycle, the energy from the electron carriers is used in carbon fixation, the conversion of carbon dioxide and water into carbohydrates. RpiA is essential in the cycle, as Ru5P generated from R5P is subsequently converted to ribulose-1,5-bisphosphate (RuBP), the acceptor of carbon dioxide in the first dark reaction of photosynthesis (Figure 3). The direct product of RuBP carboxylase reaction is glyceraldehyde-3-phosphate; these are subsequently used to make larger carbohydrates. Glyceraldehyde-3-phosphate is converted to glucose which is later converted by the plant to storage forms (e.g., starch or cellulose) or used for energy.
Protein Genetics
RpiA in human beings is encoded on the second chromosome on the short arm (p arm) at position 11.2. Its encoding sequence is nearly 60,000 base pairs long. The only known naturally occurring genetic mutation results in ribose-5-phosphate isomerase deficiency, discussed below. The enzyme is thought to have been present for most of evolutionary history. Knock-out experiments conducted on the genes of various species meant to encode RpiA have indicated similar conserved residues and structural motiffs, indicating ancient origins of the gene.
RPI Deficiency
Ribose-5-phosphate isomerase deficiency is mutated in a rare disorder, Ribose-5-phosphate isomerase deficiency. The disease has only one known affected patient, diagnosed in 1999. It has been found to be caused by a combination of two mutations. The first is an insertion of a premature stop codon into the gene encoding the isomerase, and the second is a missense mutation. The molecular pathology is, as yet, unclear.
RpiA and the malaria parasite
RpiA generated attention when the enzyme was found to play an essential role in the pathogenesis of the parasite Plasmodium falciparum, the causative agent of malaria. Plasmodium cells have a critical need for a large supply of the reducing power of NADPH via PPP in order to support their rapid growth. The need for NADPH is also required to detoxify heme, the product of hemoglobin degradation. Furthermore, Plasmodium has an intense requirement for nucleic acid production to support its rapid proliferation. The R5P produced via increased pentose phosphate pathway activity is used to generate 5-phospho-D-ribose α-1-pyrophosphate (PRPP) needed for nucleic acid synthesis. It has been shown that PRPP concentrations are increased 56 fold in infected erythrocytes compared with uninfected erythrocytes. Hence, designing drugs that target RpiA in Plasmodium falciparum could have therapeutic potential for patients that suffer from malaria.
Structural studies
As of late 2007, 15 structures have been solved for this class of enzymes, with PDB accession codes 1LK5, 1LK7, 1LKZ, 1M0S, 1NN4, 1O1X, 1O8B, 1UJ4, 1UJ5, 1UJ6, 1USL, 1XTZ, 2BES, 2BET, and 2F8M.
References
- Dickensen F., Williamson D. H. (1956). "Pentose phosphate isomerase and epimerase from animal tissues". Biochem. J. 64 (3): 567â€"78. PMC 1199776. PMID 13373810.Â
- Horecker B. L., Smyrniotis P. Z., Seegmiller J. E. (1951). "The enzymatic conversion of 6-phosphogluconate to ribulose-5-phosphate and ribose-5-phosphate". J. Biol. Chem. 193 (1): 383â€"96. PMID 14907726.Â
- Hurwitz J., Weissenbach A., Horecker B. L., Smyrniotis P. Z. (1956). "Spinach phosphoribulokinase". J. Biol. Chem. 218 (2): 769â€"83. PMID 13295229.Â







Stereoisomers with the same ordering of individual bonds and the same connection type differ in the three-dimensional arrangement of bonded atoms. Intramolecular lyases, isomerase introduction
ReplyDelete